Abbreviations
SGR : Stay-Green ; Zj: Zoysia japonica; Zmw: Zoysia matrella; Zp: Zoysia pacifica; Chl: Chlorophyll; PAO: Pheophorbide a oxygenase; BLAST: Basic Local Alignment Search Tool; CDS: Coding Sequence; SNP: Single Nucleotide Polymorphism; MeJA: Methyl jasmonic acid; ABA: Abscisic acid.
Introduction
Turfgrasses are known to reduce the amount of anthropogenic pollutants in the atmosphere (Lal, 2009). Their use in urban areas and towns can help reduce carbon dioxide emissions, energy use and other factors that contribute to the fight against climate change. Given all the advantages of turfgrasses, there has not been much research on the effect of different abiotic stresses on the commonly cultivated turfgrasses.
Depending on the species and stage of development of the plant, several coping mechanisms are employed by plants to deal with abiotic stress (Mickelbart et al., 2015). The strategy employed by plants to escape the stress involves accelerated senescence and leaf abscission, reduces their visual quality and biomass accumulation (Kooyers, 2015). The most noticeable sign of senescence is a change of leaf color, which often shifts from green to yellow or red, and ultimately brown (Koyama, 2018). Due to the fact that the market value of turfgrasses is typically decided by visual quality, the study of the genetic mechanism of senescence is of special importance. Chloroplast disintegration is one of the key steps in senescence. The degradation of chlorophyll (Chl) during senescence occurs in a multistep Pheophorbide a oxygenase (PAO) pathway. Another key regulator of Chl degradation is the STAY-GREEN (SGR) proteins that work independently of PAO pathway (Hörtensteiner, 2006). Many studies have shown that the overexpression of the SGR gene induces chlorophyll breakdown, indicating that this protein is probably a master regulator of Chl degradation. Thomas and Smart have described Stay-green mutants as plants with leaves that remain green longer as compared to their parental genotypes (Thomas and Smart, 1993). Five types of SGR mutants have been identified (A-E), among them, the type C mutant shows delayed degradation of chlorophyll and a normal declined rate of photosynthesis. Type C mutants are also known as cosmetic stay-greens because phenotypically, they remain green during senescence.
Zoysiagrasses (Zoysia spp.) are commonly used because of their higher adaptability to variable temperatures and their shade tolerant properties (Patton, 2009). Z. japonica SGR gene has been characterized and its overexpression in Arabidopsis resulted in rapid chlorophyll degradation and decomposition of chloroplast structures (Teng et al., 2016). In the present study, we predicted the SGR genes in Z. matrella and Z. pacifica using bioinformatic tools. Two SGRs were predicted in Z. matrella and three in Z. pacifica. We report the analysis of the gene structure, the distribution of the transcription regulatory elements, the phylogenetic relationship of the six SGR genes and their protein structure prediction. This study aims to understand the effect of genetic variation in the different SGRs in the three Zoysiagrasses using comparative genomics approaches.
Materials and methods
Identification and sequence data retrieval of Zoysia Stay-Green (SGR) genes
The full coding sequence and genomic DNA sequence of the stay-green gene from Zoysia japonica cultivar “Zenith” (ZjSGR) was obtained from the NCBI database. The genomic sequences of Zoysia japonica (Zj), Zoysia matrella (Zmw), and Zoysia pacifica (Zp) were downloaded from the ‘Zoysia Genome Database’(). To identify SGR genes in Z. pacifica and Z. matrella, a BLAST search using the ZjSGR sequence as query was done using the following parameters: expected values ≤1E-5 and more than 80 percent coverage. After retrieving all of the BLAST results, the NCBI Conserved Domain database was used to conduct a search for conserved domains. The construction of intron-exon of the predicted SGR genes was constructed by manual alignment of the CDS with the corresponding genomic sequences. SNP-sites were used to identify SNPs and in/dels (Page et al., 2016). The T-COFFEE multiple sequence alignment server was used to perform multiple alignments of the putative SGR genes (Di Tommaso et al., 2011). Phylogenetic tree was constructed with MEGA X (https://www.megasoftware.net/), using maximum likelihood method with 1,000 bootstrap replications.
Identification of Zoysia SGR upstream regulatory region (URR)
A 2 kb sequence upstream of the transcription start site was extracted to identify the upstream regulatory domains of the Zoysia SGR genes. Transcription Start Sites Plant (TSSP) was used to estimate the probable promoter and TATA box sites (Shahmuradov et al., 2017). The transcription binding sites in the URR were predicted using PlantTFDB (Jin et al., 2016).
Protein structure prediction
The physical and chemical parameters of the predicted SGR proteins in the three Zoysiagrasses was computed using the ProtParam tool from Expasy (Gasteiger et al., 2005). Using Protein Homology/analogY Recognition Engine 2 (PHYRE2), the tertiary structures of the SGR proteins were predicted and Chimera 1.15 was used for visualization of the protein structures (Kelley et al., 2015). Using fPocket2 (http://fpocket.sourceforge.net/), active ligand binding sites of the proteins were detected.
Results and Discussion
Identification and characterization of the SGR genes in the three Zoysiagrasses
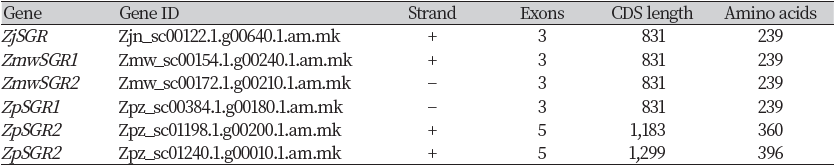
The stay-green gene (ZjSGR) in Zoysia japonica cultivar “Zenith” has been previously isolated and its role in chlorophyll degradation has been published (Teng et al., 2016). The study revealed that ZjSGR was induced by darkness, MeJA and ABA and its expression level was upregulated by natural senescence. Using this information, we obtained the homologous genes in Z. matrella and Z. pacifica. Two BLAST hits were obtained in Z. matrella genome, and three hits in Z. pacifica genome (Table 1). Both Z. matrella SGR genes (ZmwSGR1,2) and one Z. pacifica SGR gene (ZpSGR1) comprised of three exons and two introns, with an open reading frame of 831 bases encoding 239 amino acids (Table 1) (Fig. 1). The two Z. pacifica SGR genes; ZpSGR2 and ZpSGR3 consisted of 5 exons and 4 introns each (Fig. 1). The ORF in ZpSGR2 comprises of 1,183 bases, coding for 360 amino acids, whereas in ZpSGR3, the ORF of 1,299 bases encodes 396 amino acids (Table 1). In Arabidopsis thaliana, two stay-green genes have been reported; AT4G22920 (AtSGR1) and AT4G11910 (AtSGR2), consisting of 4 exons (Aubry et al., 2008). The SGR gene in the tall fescue grass, FaNYE1, consisted of three exons and two intronic regions (Wei et al., 2011).
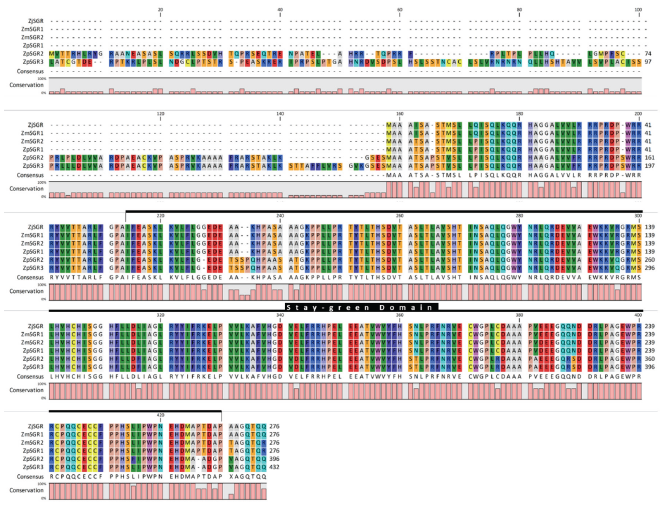
Multiple sequence alignment of the SGR coding sequences in the three Zoysiagrasses revealed that the ZmwSGR1 coding sequence (CDS) was identical to the ZjSGR. The SNP analysis of the predicted SGR CDS was done using ZjSGR as a reference. The highest amount of SNP sites were seen in ZpSGR2 (56), followed by ZpSGR3 (54), ZpSGR1 (11) and ZmwSGR2. ZpSGR2 had a 345 bp long insertion and ZpSGR3 had a 462 bp insertion at the start of the CDS. In addition, both ZpSGR2 and ZpSGR3 had 3 bp insertion sites and 3 bp deletion sites each. Conserved domain analysis of the amino acid sequence of the SGR proteins revealed a single domain belonging to the Stay-green superfamily in all the three Zoysiagrasses (Fig. 2). The results of sequence comparisons in previous studies have shown that SGRs are highly conserved proteins without any characterized domains, which supports the idea that SGRs encode regulatory proteins that control chlorophyll breakdown rather than acting as a specific enzyme (Hörtensteiner, 2006; Park et al., 2007). Using the maximum likelihood method, the phylogenetic relation among the six SGR genes was determined. The ZpSGR2 and ZpSGR3 formed a distinct group A and the remaining SGR genes were grouped together in group B (Fig. 1). Group B was further divided into subgroup C containing ZjSGR and ZmwSGR1; and subgroup D containing ZpSGR1 and ZmwSGR2.
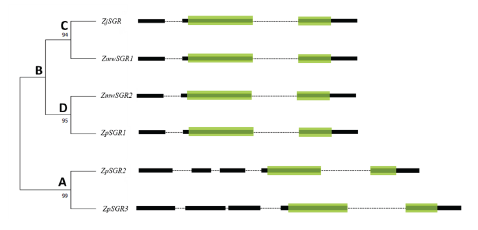
Fig. 1. Phylogenetic relationship of SGR genes among the three Zoysiagrasses using maximum likelihood method with 1,000 bootstrap replications. The green boxes represent the Stay-green domain, black boxes represent exons and dotted lines represent introns. A and B denotes major groups, and C-D denotes subgroups.
Zoysia SGR upstream regulatory region analysis and identification of transcription factor binding sites
By interacting with certain upstream regulatory regions (URR), transcription factors (TFs) can temporally and spatially switch on or off the transcription of their target genes. The 2 kb sequence upstream of the transcription start site was extracted to predict the promoter and TATA box. All the SGR had two promoters and an accompanying TATA box except for ZpSGR2 and ZpSGR3 (Fig. 3A). Additionally, ZjSGR had a predicted enhancer at position -476, and ZmwSGR2 had a third promoter and TATA box set predicted. The positions of the predicted promoters were highly conserved among the Zoysiagrasses.
Analysis of the transcription factor binding sites in the upstream regulatory regions revealed the presence motif belongs to the following families: BBR-BPC, C2H2, Dof, HD-ZIP, MIKC_MADS, MYB, Trihelix, NAC, WRKY, YABBY and ZF-HD (Fig. 3B). The main transcription factors that have been reported to take part in leaf senescence belong to NAC (Guo and Gan, 2006; Kim et al., 2009), WRKY (Zhang et al., 2016), AP2 (Phukan et al., 2017) and MYB (Zhang et al., 2012) families. One of the biggest families of plant TFs is the NAC (NAM, ATAF, and CUC) gene family. During natural leaf senescence their enhanced expression level has been reported in Arabidopsis (Breeze et al., 2011). In our analysis the NAC motifs were present only in ZmwSGR2 and ZpSGR2 (three motifs each) (Fig. 3B). All the Zoysia SGRs had MYB or MYB related binding motifs in the upstream regulatory regions. The transcriptional analysis in barley and maize showed an association of MYB TFs with leaf senescence and nitrogen remobilization (Hollmann et al., 2014; Zhang et al., 2014). In rice, OsMYB102 showed association with leaf senescence by down-regulating the abscisic acid accumulation and signaling (Piao et al., 2019). WRKY TF binding motifs were only present in ZpSGR2 and ZPSGR3. The regulatory mechanisms or several WRKY genes during senescence have been well documented in Arabidopsis (Guo et al., 2017).
Protein structure prediction and the analysis of the Stay-green domain
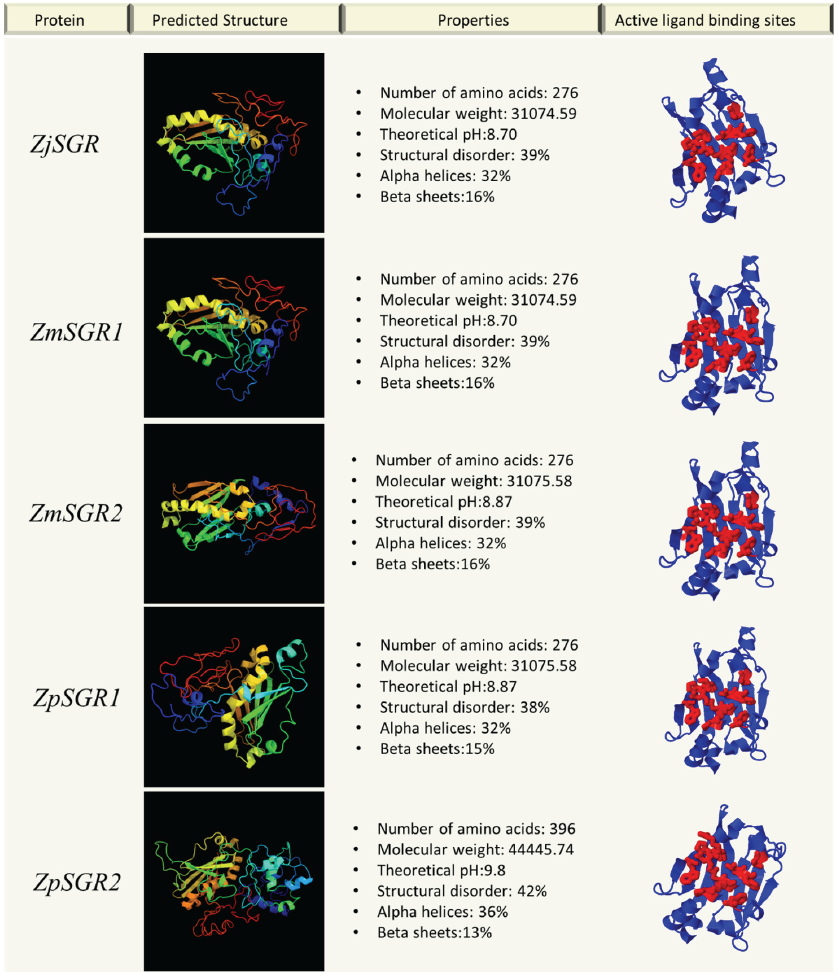
The physical and chemical properties of the predicted genes were analyzed. The properties of the ZjSGR, ZmwSGR1, ZmwSGR2 and ZpSGR1 were nearly similar (Fig. 4). In ZpSGR2 and ZpSGR3, the pH was higher than the other Zoysiagrasses and the structures were more disordered. ZpSGR2 protein structure comprised of 36% alpha helices and 13% beta sheets, whereas ZpSGR3 comprised of 26% alpha helices and 19% beta sheets. The analysis of single amino acid polymorphisms (SAP) in the Stay-green domain revealed one SAP each in ZmwSGR1 and ZpSGR1, and fourteen and thirteen SAP sites in ZpSGR1 and ZpSGR2 respectively. There were two amino acid insertion sites and one deletion site seen in ZpSGR2 and ZpSGR3. The effect of amino acid substitutions was seen in the predicted structures of the Zoysia SGR proteins. Protein pocket detection of the predicted proteins resulted in an identical ligand binding site in all the Zoysia SGRs, except ZpSGR3 (Fig. 4).
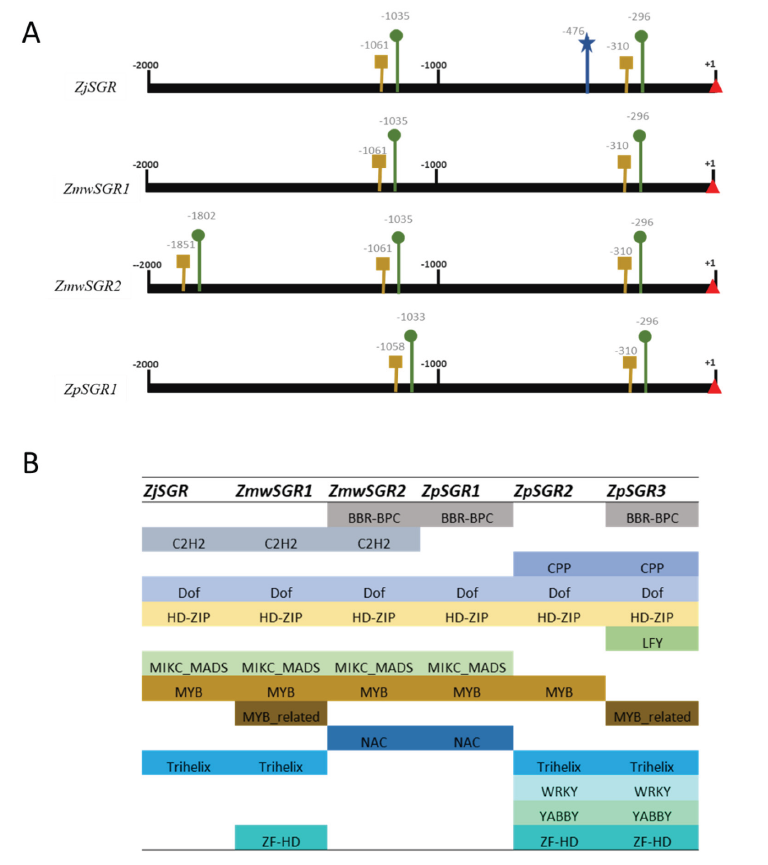
Fig. 3. Analysis of the upstream regulatory region of the Stay-green genes (SGR) predicted in the three Zoysiagrasses. A) The predicted promoter and TATA box sites are in denoted in green and yellow, respectively. Transcription start site (+1) is marked by red triangle. Predicted enhancer sites have been marked in blue. No promoters were predicted in ZpSGR2 and ZpSGR3. B) Transcription factor families identified in the upstream regulatory region of the Zoysia SGR genes.
Conclusion
The impact of climate change on biological systems is occurring at a higher rate than in the past decades. Its effect includes rising temperatures, more erratic precipitation patterns, and a rise in atmospheric CO2. Its implications have been well studied in primary agricultural crops, but there have not been many studies in turfgrasses, which are an essential part of the ecosystem. Their preservation and development will add to enhancing the ecosystem and social values. Zoysiagrasses have been widely used in landscaping. Each Zoysiagrass (Z. japonica, Z. matrella and Z. pacifica) has its own geographical drawback. For example, Z. japonica has the highest cold tolerance but cannot grow well in saline soil and drought affected areas. Z. matrella has good salt and shade tolerance but poor cold tolerance. Whereas Z. pacifica is a high quality turfgrass with poor cold tolerance but excellent shade tolerance (Patton and Reicher, 2007; Unruh et al., 2011). All these abiotic stresses ultimately result in leaf browning, thereby affecting the appearance of lawns. We elucidated five SGR genes, two in Z. matrella and three in Z. pacifica and compared them to reveal differences in the final protein structures which may provide some insight into the differences in the senescence induced chlorophyll degradation and the underlying phytohormonal communication. To compare the effect of the genetic variations identified in this study, further experimentation is required. The data presented in this study attempts to supplement the otherwise limited Zoysiagrass research pool.