Introduction
Zoysiagrasses are warm season turfgrasses. These comprise of eleven allotetraploid species that possess varied morphological and physiological traits. Many members of the Zoysia species are cold and salt tolerant among the warm-season turfgrasses (Magni et al., 2017). Turfgrasses from the Zoysia genus are also hardy, shade resistant and are preferred for their athletic characteristics (Wherley et al., 2011). While zoysiagrass research lags behind other members of the Poaceae family, recent genetic studies are shedding light on their diversity and evolutionary history. (Tanaka et al., 2016).
Plant hormone gibberellic acid (GA) plays a critical role in various stages of development, including seed germination, stem elongation, flowering time, and flower formation (Castro-Camba et al., 2022). Plants with mutations affecting GA production or signaling often show stunted growth, delayed flowering, or complete failure to flower (Sun, 2008). DELLA proteins function as repressors, hindering GA-dependent growth responses. However, GA triggers the degradation of these DELLA proteins, increasing growth. Recent research suggests that DELLA activity, and other components of GA signaling, are further regulated by modifications occurring after protein synthesis (post-translational modifications). Additionally, crosstalk between GA signaling and other signaling pathways within the plant appears to be important, potentially influencing GA's effects by regulating it’s signaling components.
Negative regulators of gibberellin (GA) signaling have been identified through cloning efforts in various plant species. In the model plant Arabidopsis, the gibberellin-insensitive (GAI) gene was isolated and characterized as a transcription factor that acts as a negative regulator of GA responses. Importantly, a specific mutation in GAI – a 51-bp deletion within the highly conserved N-terminal DELLA domain (Silverstone et al., 1998)– resulted in a dominant gain-of-function mutant (gai) with a dwarf phenotype due to reduced GA responsiveness (Peng et al., 1997). Intriguingly, the "green revolution" genes associated with dwarfism in cereal crops – Reduced height (Rht) in wheat, Dwarf8 (D8) in maize, Slender1 (SLN1) in barley, and Slender1 (SLN1) in rice – all represent functional orthologs of Arabidopsis GAI(Chandler et al., 2002; Ikeda et al., 2001; Peng et al., 1999) . Notably, similar to the Arabidopsis gai mutant, deletions within the DELLA domain of these GAI orthologs consistently lead to dominant dwarf phenotypes.
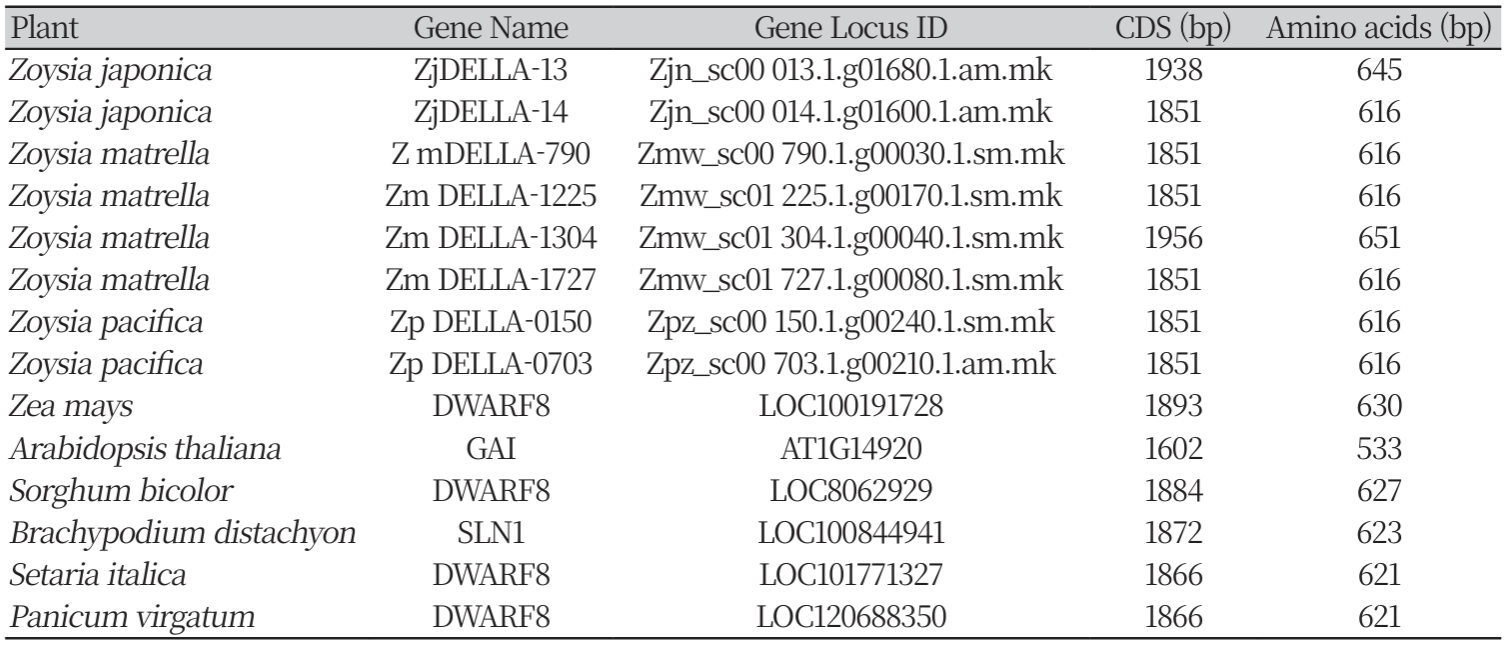
In the present study, we predicted the DELLA genes in zoysiagrasses like the DWARF8 gene from Zea mays using bioinformatic tools. We identified eight putative DELLA protein sequences within the zoysiagrass genomes. This study aims to understand the effect of genetic variation in the different Zea mays DWARF8 orthologs in the three zoysiagrasses using comparative genomics approaches. The anticipated results will give understanding of the difference in the plant height and mowing frequency among the three zoysiagrasses.
Materials and methods
Identification and sequence data retrieval of DELLA proteins
In order to search for the DELLA proteins similar to the DWARF8, The Zea mays DWARF8 gene sequence (NCBI Gene ID: 100191728) was retrieved and used as query to perform the BLAST search on the Zoysia Genome Browser (https://zoysia. kazusa.or.jp/index.html). Highly similar sequences to Z. mays D8 with E value 0.0, were retrieved from Zoysia japonica, Zoysia matrella, and Zoysia pacifica Additionally, DWARF8 orthologs in closely related species; Sorghum bicolor, Setaria italica, Panicum virgatum and Brachypodium distachyon were retrieved from NCBI. The sequence of the Arabidopsis GAI (a known ortholog of ZmDWARF8), was retrieved from TAIR.
Phylogenetic analysis
The amino acid sequences of all the plants used in this study were aligned using default parameters in Clustal Omega (https://www. ebi.ac.uk/jdispatcher/msa/clustalo). The aligned sequences were used to construct an evolutionary tree with MEGA11 software using maximum likelihood method using 1000 bootstrap replications. The phylogenetic tree was uploaded to the iTOL webserver (http:// itol.embl.de/) in Newick format for better visualization.
Gene structure assessment and conserved domain analysis
The gene structure of the predicted DELLA proteins of the three zoysiagrasses, and the six plant species were predicted using Gene Structure Display Server 2.0 (https://gsds.gao-lab.org/). The protein sequences of all the plants were subjected to a conserved domain search using NCBI Conserved Domain Database (https://www.ncbi.nlm.nih.gov/cdd/).
Identification of cis-regulatory elements
For cis -element analysis, the 1500bp sequence upstream of the transcription start site was extracted in the predicted DELLA proteins. The probable promoter region was identified using Transcription Start Sites Plant (TSSP). Using Plant Transcription factor database (PlantTFDB), the transcription factor binding sites in the upstream regulatory region were predicted.
Protein sequence analysis and structure prediction
The physical and chemical parameters of the predicted SGR proteins in the three zoysiagrasses were computed using the ProtParam tool from Expasy (Gasteiger et al., 2005). Using PSIPRED and SWISS-MODEL, the three-dimensional protein structures were predicted, analyzed, and visualized.
Results and Discussion
Identification and characterization of the predicted DELLAs
We identified eight DELLA proteins using the known Z. mays DWARF8 peptide sequence as query and performed BLASTP search on the Zoysia genome database. Of these eight DELLA proteins, two belonged to Z. japonica, four to Z. matrella and two to Z. pacifica (Table 1). The proteins identified were named based on their loci for ease of reference. The coding sequence of the eight zoysia DELLA proteins ranged from 1851bp to 1956bp. These species were analyzed due to their close relationship with the zoysiagrasses. ZmDWARF8 and AtGAI are well studied and have been functionally characterized (Andersen et al., 2005; Peng et al., 1997). The BLASTP hits corresponding to ZmDWARF8 (Camus-Kulandaivelu et al., 2008) resulted in DWARF8 in Setaria italica, Sorghum bicolor, and Panicum virgatum, and SLN1 (SLENDER1) in Brachypodium distachyon . The DWARF8 orthologs in the above species have limited information, and the sequences downloaded from NCBI have been predicted by automated computational analysis using NCBI’s gene prediction method called Gnomon, and supported by EST evidence (Souvorov et al., 2010). The information of all the DWARF8 genes used in this study has been provided in Table 1.
Evolutionary relationship and gene structural analysis of the predicted DELLA genes
The evolutionary history among the Z. mays DWARF8, AtGAI, and the predicted DELLA proteins in zoysiagrasses and related species was deduced using the maximum likelihood method with 1000 bootstrap replications. This revealed that the DELLA proteins originated from a common ancestor and diverged into two groups; one comprising of the zoysiagrass predicted DELLA proteins and the other with Z. mays and other members of the Poaceae family. The predicted DELLA proteins in the zoysiagrasses formed four closely related subgroups, while S. bicolor, S. italica, P. virgatum, and B. distachyon DELLA proteins were grouped with Z. mays DWARF8. ZmDWARF8 and AtGAI genes have one exon each, constituting an N-terminal DELLA domain and a C-terminal GRAS domain (Huang and Yu, 2009; Lawit et al., 2010). The structures of the corresponding DELLA genes predicted in the three zoysiagrasses and related plant species were identified on Gene Structure Display (GSDS), by comparing the genomic sequences and the corresponding coding sequences of the DELLA genes. The DELLA genes; ZjnDELLA-13 and ZmwDELLA-1304 constituted two exons, while all the other DELLA genes in the zoysiagrasses and related species consisted of one exon. Conserved domain analysis revealed that all the predicted DELLA genes constituted an N-terminal DELLA domain and a C-terminal GRAS domain. Multiple sequence alignment of the predicted DELLA proteins, ZmDWARF8 and AtGAI revealed a high degree of conservation in the DELLA domain and GRAS domains of each subgroup, suggesting a similar evolutionary relationship.
Promoter region identification and cis-element analysis in the predicted DELLA genes
Previous research has shown that DELLAs play a role in various plant hormone signaling pathways by interacting with different transcriptional regulators and factors. However, the exact way DELLAs interact with these regulators and how they are themselves controlled is not fully understood. In this study, we investigated the potential functions and regulatory mechanisms of the predicted DELLAs in the three zoysiagrasses and the four related plant species. We analyzed the 1kb base pair region upstream of the predicted DELLA genes, focusing on regulatory elements. We used the TSSP database to identify the probable promoter region (Figure 3). All the predicted DELLA genes had at least one promoter except for ZmwDELLA-1304. At least one TATA-/ enhancer was predicted for each gene. Analysis of the transcription factor binding sites in the upstream regulatory regions revealed the presence of the following major transcription factor families: TALE, ERF, C2H2, MYB, HD-ZIP, Dof, WOX, SBP, HSF, bZIP, NAC, bHLH, Trihelix. ERF/AP2 transcription factors are known to interact with DELLA proteins in Arabidopsis (Feng et al., 2020; la Rosa et al., 2014) . ERF TFs were identified in the URR of zoysia DELLA genes of ZmwDELLA-1727, ZjnDELLA14, ZmwDELLA790, ZpwDELLA150, ZpzDELLA-703 and Zmw1225. No ERF binding sites were identified in B. distachyon SLN1. MYB transcription factors are the largest transcription factor families, known to play a diverse role in multiple biological pathways such as plant growth and development. MYBs also play a role in GA pathway by interacting with DELLA proteins to modulate gibberellin and jasmonate signaling (Qi et al., 2014). MYB TF binding sites were identified in the URR of B. distachyon, ZmwDELLA-1727, ZjnDELLA-14, ZmwDELLA-790, and ZpzDELLA-150. Beyond the structural variations, further studies are needed to verify different TFs that modulate the functional difference of zoysia DELLAs among three species.
Protein sequence analysis and secondary structure prediction DELLA proteins
The physical and chemical properties of the predicted DELLA proteins were analyzed. An in-silico analysis of the amino acids in the DELLA protein’s secondary structure was performed using SWISS-model. All the zoysiagrass predicted DELLA proteins were found to match the AlphaFold DB model of the DELLA protein DWARF8 in Zea mays to about 91% identity (Figure 4). The protein secondary structures were highly similar to each other. The theoretical PI ranged between 4.87 to 5.11. The protein modelling resulted in monomers and no ligands were identified.
Fig. 4
Physical, chemical, and structural analysis of eight Zoysia DELLA proteins. For each DELLA protein, the secondary structure was predicted using SWISS-model (image colored by rainbow N to C terminus). The chemical and physical properties of each protein were predicted using the ProtParam tool on Expasy. All the structures were modeled based on the AlphaFold model of Zea mays DWARF8 as a template.