Abstract
Prickly lettuce (
Figures & Tables
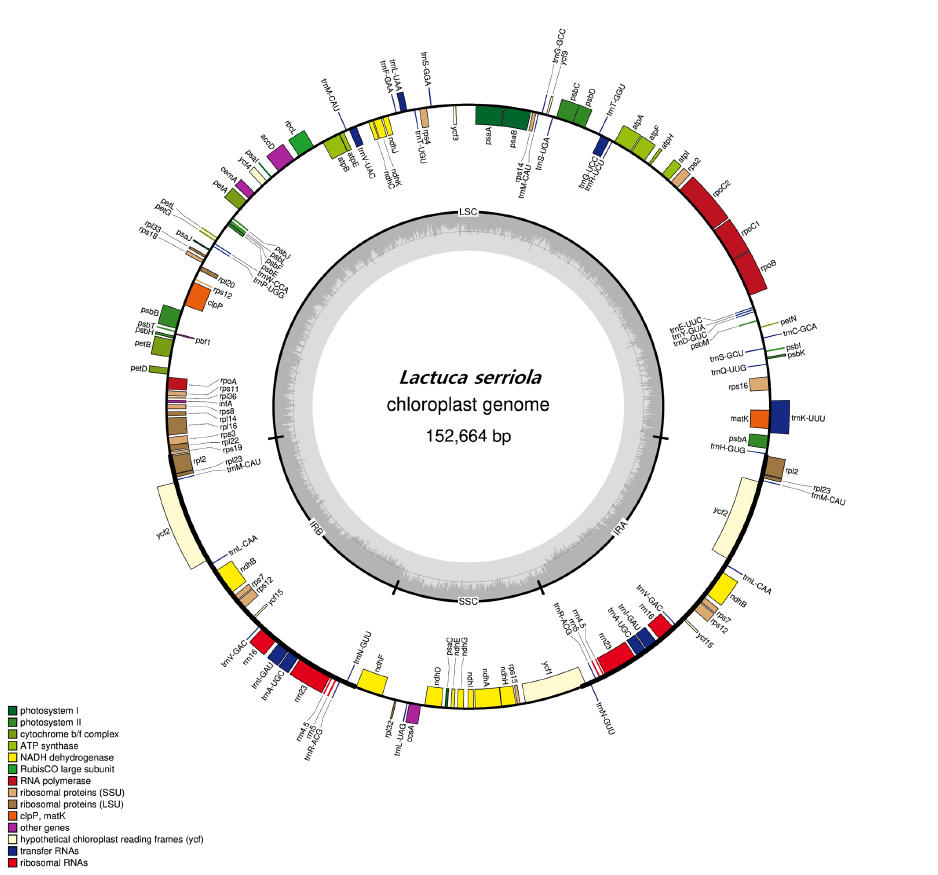
Fig. 1.Circular genome map of chloroplast of .
Gyeong-Jong Heo1 Jin-Won Kim1,*
1Crop Protection Division, National Institute of Agricultural Sciences, RDA, Wanju 55365, Korea
Prickly lettuce (

Fig. 1.Circular genome map of chloroplast of .