Abstract
Senescence is a response regulator to abiotic and biotic stress for plants, as well as an important developmental process. Plant senescence and the breakdown of chlorophyll depend on the stay-green proteins. Chlorophyll degradation during abiotic stresses is an undesirable trait in grasses that are mainly grown for adding greenery in the landscaping industry. The underlying processes of stay-green gene were poorly researched in the
Figures & Tables
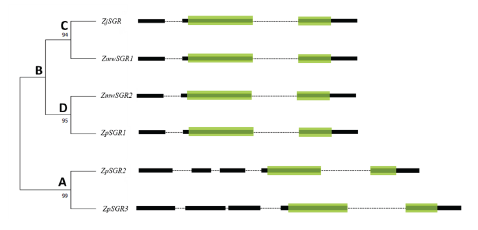
Fig. 1. Phylogenetic relationship of SGR genes among the three grasses using maximum likelihood method with 1,000 bootstrap replications. The green boxes represent the Stay-green domain, black boxes represent exons and dotted lines represent introns. A and B denotes major groups, and C-D denotes subgroups.


