Introduction
Zoysiagrasses have been used widely in lawns because they are warm-season turf and environment-friendly. Being perennial, it also becomes one of the limiting factors while growing them in transitional and temperate regions, due to its low tolerance to cold stress. Cold stress has a strong impact on the plants, it largely reduces the activity of particularly the enzymes involved in photosynthesis and destabilizes the cell membrane (Thakur and Nayyar, 2013). A plant’s ability to withstand low temperatures (0-15℃) without injury or damage is referred to as chilling tolerance, whereas the increased tolerance of the plant to minimize the damage caused by low temperatures is termed cold acclimatization (Thomashow, 1999). Both the processes involve a cascade of biochemical, molecular, and metabolic processes (Thakur and Nayyar, 2013).
There are three main cold-responsive genes in plants: Inducer of CBF expression (ICE), C-repeat Binding Factors (CBFs), and Cold-regulated genes (CORs). These three key players constitute a cold-responsive pathway called the ICE-CBF-COR cascade that alleviates cold stress in plants. In Arabidopsis, two ICE genes have been documented: ICE1 and ICE2 which encode MYC-type bHLH transcription factors in response to cold temperature. Both ICE genes regulate cold stress response and stomatal development by activating the expression of CBF3 under cold stress (Hwarari et al., 2022; Kurbidaeva et al., 2014). While ICE1 confers freezing tolerance to the whole plant, ICE2 was seen to be functionally specific to meristems in Arabidopsis. Bioinformatic analyses revealed that there was sequence diversification of ICE2 from ICE1 and the gain of cis-acting elements in the ICE2 promoter may be the reason behind its expression specificity to meristem (Kurbidaeva et al., 2014).
Cold tolerance in zoysiagrass species varies with Z. japonica being the most cold-tolerant than Z. matrella, and Z. pacifica being the least cold-tolerant (Patton and Reicher, 2007). Cold tolerance and its molecular mechanism have been widely studied in Arabidopsis and sparsely in Z. japonica. However, the underlying genetic mechanism and the differences in the ability to thrive in cold have not been studied well in Z. matrella and Z. pacifica. The objectives of this research were (i) to predict ICE1 and ICE2 genes in Z. matrella and Z. pacifica, (ii) to compare the genetic sequence variation in the predicted ICE1 and ICE2 among the zoysiagrasses, and (iii) to identify the probable effect of the ICE1 and ICE2 sequence variation on the cold tolerance properties of the zoysiagrasses.
Materials and methods
Sequence data retrieval and identification of ICE1/2 genes in zoysiagrass
Zoysia japonica ‘Nagirizaki’ (Zjn), Zoysia matrella ‘Wakaba’ (Zmw), and Zoysia pacifica ‘Zanpa’ (Zpz) genome sequences were downloaded from ‘Zoysia genome database’ (). The full coding sequence and protein sequence of Arabidopsis and Z. japonica ICE1 and ICE2 were obtained from TAIR () and NCBI (), respectively. These sequences were used as a query sequence for a BLAST (Basic Local Alignment Search Tool) search against Z. pacifica and Z. matrella genome with the following parameters: expected values ≤1E-5 and more than 80 percent coverage. Single nucleotide polymorphisms (SNPs) and In/Dels were identified using SNP-sites (Page et al., 2016). All the BLAST hits were retrieved, and a conserved domain search was performed using NCBI conserved domain search. Multiple alignments of the putative ICE1 and ICE2 genes were done using ClustalW (Thompson et al., 1994) with the default settings.
Identification of ICE1, 2 promoter region
The promoter regions of the Zoysia ICE1, 2 genes were identified by extracting a 1kb sequence upstream of the transcription start site. The putative promoter and TATA box sites were predicted using TSSP (Shahmuradov et al., 2017). Further, PlantRegMap (Jin et al., 2016) was used to identify transcription factor binding sites upstream of the predicted ICE1, 2 transcription start sites.
Results
Identification and characterization of Zoysia ICE1 and ICE2 genes
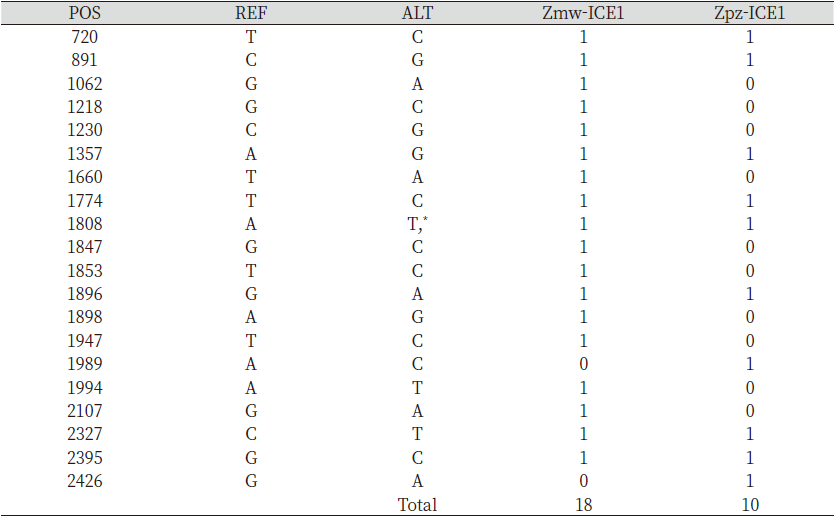
ICE1 and ICE2 have been known to have a freezing protective role in Arabidopsis. these basic-helix-loop helix transcription factors have been widely used to confer cold tolerance in transgenic plants. The information regarding Zoysia japonica ICE1 (Zjn-ICE1) and ICE2 (Zjn-ICE2) full-length coding sequences has already been published. Using Zoysia japonica ICE1 and ICE2 full-length coding sequences as the BLAST query, the predicted ICE1 and ICE2 gene sequences in Zmw and Zpz genome were obtained. The Zjn-ICE1 coding sequence constitutes 2,451 bp, whereas the predicted Zmw-ICE1 and Zpz-ICE1 constituted 2,421 and 2,436 bp, respectively (Table 1).
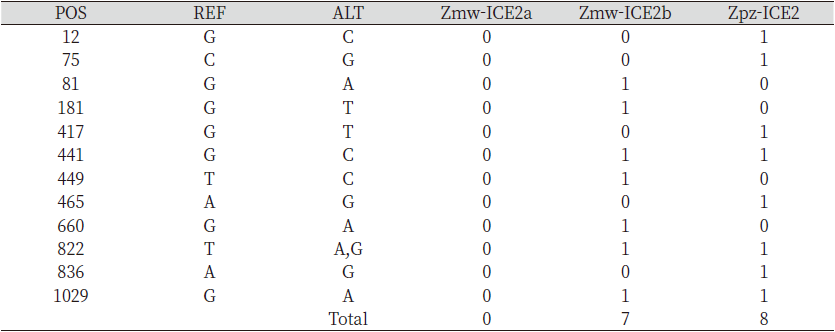
The analysis of single nucleotide polymorphic sites (SNPs) in the predicted Zmw-ICE1 and Zpz-ICE1 with respect to the Zjn-ICE1 coding sequence revealed 18 and 11 SNPs respectively, Zmw-ICE1 and Zpz-ICE1 had 30 bp and 37 bp deletions with respect to Zjn-ICE1 (Table 2). Additionally, Zpz-ICE1 had a 22 bp insertion compared to Zjn-ICE1. Interestingly, ICE2 gene sequence retrieval resulted in two hits in the Z. matrella genome with respect to Z. japonica, which was termed as Zmw-ICE2a and Zmw-ICE2b, respectively. These two predicted sequences were identical in length (1,110 bp) except for 7 SNPs in the coding sequence with respect to each other (Table 1). The Zmw-ICE2a sequence was identical to that of Zjn-ICE2, whereas Zmw-ICE2b had 7 SNPs with respect to Zjn-ICE2 (Table 3). The ICE2 coding sequence predicted in Z. pacifica (Zpz-ICE2), had a 3 bp insertion, and 9 SNPs as compared to the Zjn-ICE2 (Table 3).
ICE1, 2 protein domain conservation
Inducer of CBF expression 1,2 (ICE1,2) transcription factors (TFs) have two distinct domains that recognize the CBF/DREB1 by identifying the MYC regulation sites (Ohta et al., 2018). The first domain, bHLH_AtAMS_like, is the basic helix-loop-helix (bHLH) domain found in Arabidopsis thaliana protein aborted microspores (AMS); a large family of transcription factors like AMS, ICE1, and SCREAM2 (Lian et al., 2017). The second domain identified belonged to the ACT domain repeat protein family, which is proposed to have a regulatory/sensory function in plants (Hsieh and Goodman, 2002). Sequence comparison revealed that all the predicted Zoysia ICE1/2 protein sequences shared the same highly conserved regions (Fig. 1). Furthermore, Zmw-ICE1 and Zpz-ICE1 comprised of a third domain; PLN02983, a biotin carboxyl carrier protein of acetyl-CoA carboxylase (Lu et al., 2020), with no known ties to the cold-responsive mechanism in plants. A closer look in the PLN02983 region showed a single amino acid polymorphism at position 453, where threonine (T) is substituted by alanine (A) in Zmw-ICE1 and Zpz-ICE1. The presence of the bHLH and ACT domain further validated the sequences of the predicted ICE1, 2 gene sequences in Z. matrella and Z. pacifica.
Table 1. List of predicted Zoysia ICE1 /2 genes.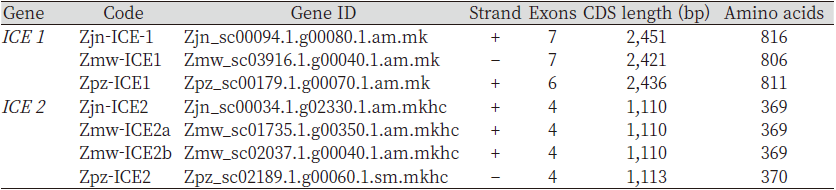
|

Fig. 1. Conserved domain analysis of the Zoysia A) Inducer of CBF expression 1 (ICE1 ) putative protein sequences and B) Inducer of CBF expression 2 (ICE2 ) putative protein sequences. Two major domains related to the ICE1 protein; bHLH_AtAMS_like (in blue) and ACT_UUR-ACR-like (in yellow) were identified in all three zoysiagrasses. An additional domain, belonging to the PLN02983 superfamily was identified in Z. matrella ICE1 (Zmw-ICE1 ) and Z. pacifica ICE1 (Zpz-ICE1 ) (highlighted in grey). The single amino acid polymorphic sites are in red, and the in/del regions have been highlighted in green. cov: Coverage; pid: Percent identity; Zjn: Z. japonica .
ICE1, 2 promoter analysis and identification of transcription factor binding sites
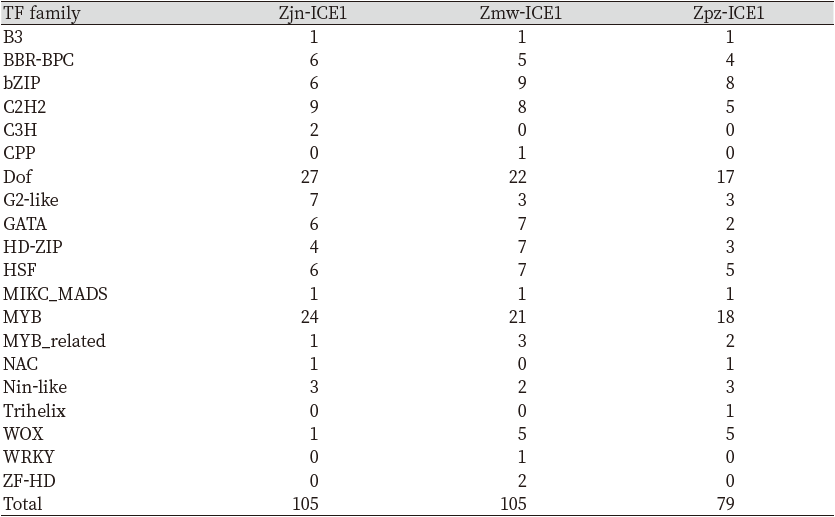
Sequences 1 kb upstream of the transcription start site were subjected to promoter site prediction. All the predicted Zoysia ICE1, 2 genes constituted at least one promoter accompanied by a TATA box (Fig. 2) The ICE1 predicted promoter region was seen to be highly conserved, whereas huge variations were seen in the sequences immediately upstream of the TSS among the Zoysiagrasses. Furthermore, there were 2 promoter and TATA box sites predicted for Zmw-ICE2a (Fig. 2). Further analyses of the probable transcription factor sites revealed 20 distinct families of TFs in Zoysia ICE1 and 22 families in Zoysia ICE2. The total number of transcription factors identified in the regulatory region of Zpz-ICE1 was 75 which was the lowest as compared to Zjn-ICE1 and Zmw-ICE1 (Table 4). MYC and Dof transcription factor binding sites were the highest in the three Zoysia ICE1 regulatory sequences. Out of the 20 transcription factor families, WRKY, ZF-D, and CPP binding sites were only found in Zmw-ICE1. Whereas C3H and Trihelix transcription factor binding sites were only seen in Zjn-ICE1 and Zpz-ICE1, respectively (Table 4).
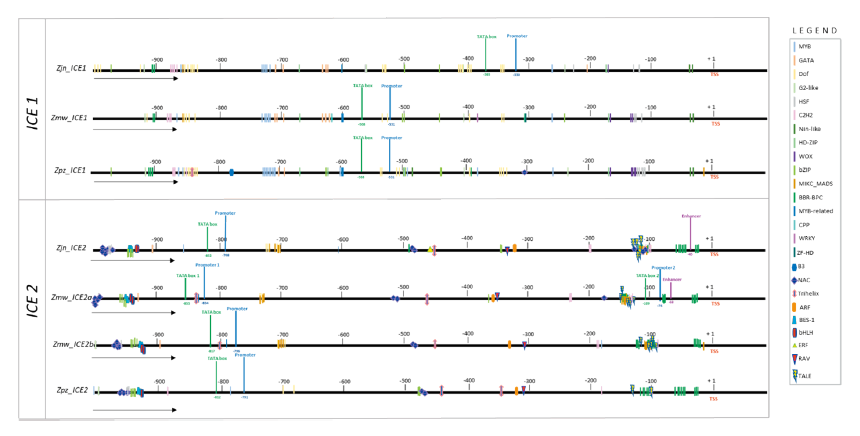
Fig. 2. Analysis of the upstream regulatory region of the Inducer of CBF expression 1, 2 (ICE1 and ICE2 ) genes in Z. japonica (Zjn), Z. matrella (Zmw) and Z. pacifica (Zpz). The predicted promoter and TATA box sites are in denoted in blue and green, respectively. The putative transcription factor binding sites have been clustered using unique symbols (see legend). The black arrows denote the direction of transcription. TSS: Transcription start site.
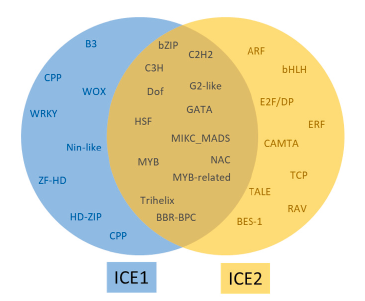
The transcription factors in the regulatory region of the ICE2 gene were highest in Zmw-ICE2a and Zmw-ICE2b, 182, and 181 respectively, whereas, Zpz-ICE2 had the least number of transcription factors (Table 5). The highest number of transcription factor binding sites in the ICE2 regulatory region of the three zoysiagrasses was occupied by the BARLEY B RECOMBINANT-BASIC PENTACYSTEINE (BBR-BPC) family followed by NAC, bZIP, and MIKC_MADS family. The ICE1 and ICE2 regulatory regions in the three zoysiagrasses have 13 transcription factors in common, and 8 TF and 9 TF families specific to only the ICE1 and ICE2 regulatory regions respectively (Fig. 3).
Discussion
Zoysiagrasses have been used as turfs all around the world due to their thick coverage and resistance to many abiotic stresses. One of the important abiotic stresses, low temperature, becomes the limiting factor for growing these grasses (Brown et al., 2020). The cold tolerance ability of the three Zoysia species; Z. japonica, Z. matrella, and Z. pacifica have been evaluated to see the relationship between leaf width and autumn growth with winter injury. It was found that Z. japonica had less winter injury than Z. matrella, followed by Z. pacifica (Patton and Reicher, 2007). However, the most commonly used zoysiagrasses turn brown on the first sign of winter in temperate regions like Korea. The genetic mechanism behind the cold tolerance abilities of each zoysiagrass has been studied to a limited extent than the other members of the Poaceae family.
This study aimed to predict the cold-responsive ICE1, 2 genes in Z. matrella and Z. pacifica and compare them with the Z. japonica ICE1, 2, which is a known positive regulator of the CBF3 gene, a key player in the cold acclimatization machinery of many plants (Wang et al., 2022). ICE genes specifically contain an MYC-like bHLH transcriptional activator that binds specifically to the MYC recognition sequences in the CBF promoter (Xie et al., 2019). The objective of this study was to predict and characterize the ICE1 and ICE2 genes in Z. matrella and Z. pacifica and to identify the probable effect of sequence variations among the ICE genes on the cold tolerance properties of the zoysiagrasses. The Zoysia Genome Database was used to identify Zmw-ICE1, 2 and Zpz-ICE1, 2 using the Z. japonica ICE1, 2 sequence. The predicted ICE1, 2 coding sequences in Z. matrella and Z. pacifica showed high sequence similarity to the Z. japonica ICE1, 2. Interestingly, two hits for the Z. matrella ICE2 were obtained, which were termed Zmw-ICE2a and Zmw-ICE2b. Zmw-ICE2a showed a hundred percent identity to the Zjn-ICE2, whereas, Zmw-ICE2b had 7 single nucleotide polymorphic sites. The ICE1 genes in Z. japonica and Z. matrella had 7 exons and Z. pacifica constituted 6 exons. The ICE2 gene in the three Zoysiagrasses constituted 4 exons each.
Conserved domain analyses revealed two major domains linked to the ICE genes: bHLH_AtAMS_like and ACT_UUR-ACR-like in both the ICE1, 2 coding sequences in all three zoysiagrasses (Hsieh and Goodman, 2002; Xie et al., 2019). The presence of an additional domain belonging to the PLN02983 superfamily was seen in Zmw-ICE1 and Zpz-ICE2, which had no probable links to cold stress tolerance in zoysiagrasses.
The regulatory regions upstream to the ICE1/2 genes were subjected to comparative analysis to identify the probable transcription factor sites. The upstream region (1 kb) from the transcription start site was compared. The regulatory regions of the ICE1, 2 genes in all the three zoysiagrasses constituted at least one promoter region and a TATA box. There were huge variations in the immediate regulatory region of the ICE1, 2 genes which may be a reason for different cold tolerance levels in the three zoysiagrasses (Das and Bansal, 2019). The diversity of transcription factor binding sites across the species contributes to the species-specific variation in gene regulation under different environmental conditions (Doniger and Fay, 2007). Further, transcription factor binding sites were analyzed in the upstream regulatory region of the ICE1, 2 genes. MYB transcription factor family constituted the highest number of binding sites in the ICE1 regulatory regions. The MYB TFs predicted in Zjn-ICE1 and Zmw-ICE1 participated in the positive regulation of transcription during the stress response, responses to salicylic acid, auxin, and abscisic acid (Park et al., 2010). The ICE2 regulatory region showed a high number of BBR-BPC transcription factor binding sites, which plays an important role in response to abiotic stresses (Ahad et al., 2021). In Arabidopsis thaliana, NAC transcription factors were shown to improve cold stress tolerance (Yuan et al., 2019). NAC transcription factor binding sites were present in high numbers in the ICE2 regulatory region than in ICE1. It may have a significant advantage on the cold stress tolerance mechanism in zoysiagrasses. Other transcription factors which are known to be involved in stress responses, like bZIP, C2H2, C3H, Dof, and trihelix were common to both the ICE1 and ICE2 regulatory regions (Table 3).
As seen in studies across the plant kingdom, the activation and deactivation of transcription factors coordinate the expression of genes in response to environmental stimuli (Lindlöf et al., 2009). There is not much information available on the abiotic stress tolerance mechanisms in the zoysiagrasses. The present study was aimed to give an informational and experimental boost in the area of abiotic stress tolerance studies in Zoysiagrasses. The predicted sequence of the Z. matrella and Z. pacifica ICE1, 2 genes can be used to identify the interactions further experimentally with CBF3 and their implications in conferring cold stress tolerance and improving the turfgrass industry. The results from this study further allow researchers to improve breeding of zoysiagrasses to withstand winter injury using molecular genetics.
Acknowledgments
This research was funded by the New Breeding Technologies Development Program (No. PJ016547), Rural Development Administration, and the Basic Science Research Program (NRF-2020R1A2C1015119) through the National Research Foundation (NRF) of Korea funded by the Ministry of Science and ICT, Republic of Korea.